Are Vision xLSTM Embedded U-Nets More Reliable in Medical 3D Image Segmentation?
Pallabi Dutta, 1 Soham Bose, 2 Swalpa Kumar Roy, 3 Sushmita Mitra 1
1 Machine Intelligence Unit, Indian Statistical Institute, Kolkata, India
2 Department of Computer Science and Engineering, Jadavpur University, Kolkata, India
3 Department of Computer Science and Engineering, Alipurduar Government Engineering and Management College, India
Abstract
The advancement of developing efficient medical image segmentation has evolved from initial dependence on Convolutional Neural Networks (CNNs) to the present investigation of hybrid models that combine CNNs with Vision Transformers. Furthermore, there is an increasing focus on creating architectures that are both high-performing in medical image segmentation tasks and computationally efficient to be deployed on systems with limited resources. Although transformers have several advantages like capturing global dependencies in the input data, they face challenges such as high computational and memory complexity. This paper investigates the integration of CNNs and Vision Extended Long Short-Term Memory (Vision-xLSTM) [1, 2] models by introducing a novel approach called UVixLSTM. The Vision-xLSTM blocks captures temporal and global relationships within the patches extracted from the CNN feature maps. The convolutional feature reconstruction path upsamples the output volume from the Vision-xLSTM blocks to produce the segmentation output. Our primary objective is to propose that Vision-xLSTM forms a reliable backbone for medical image segmentation tasks, offering excellent segmentation performance and reduced computational complexity. UVixLSTM exhibits superior performance compared to state-of-the-art networks on the publicly-available Synapse dataset. Code is available at: https://github.com/duttapallabi2907/U-VixLSTM
Highlights
- We present the first effort in studying the application of Vision-xLSTM with CNNs for medical image segmentation.
- The Vision-xLSTM blocks learn global context by modelling long-term dependencies among patches from CNN layer in the feature extraction path in a computationally effective manner as compared to the Transformer counterparts.
- UVixLSTM exhibits superior performance compared to state-of-the-art networks on the publicly-available Synapse dataset.
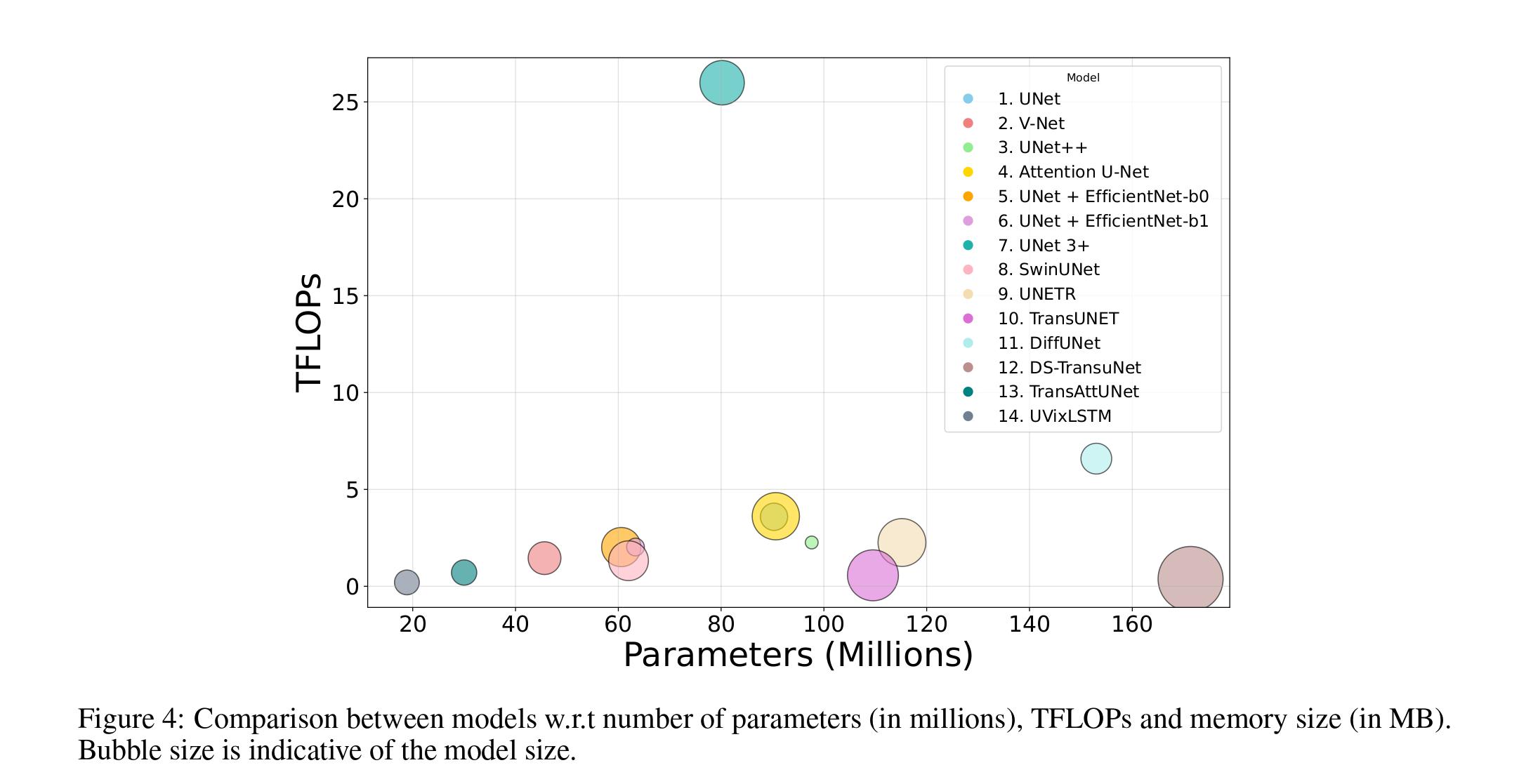
- UVixLSTM has the lowest number of parameters and TFLOPs (floating point operations per second) compared to other popular medical image segmentation algorithms.
Network
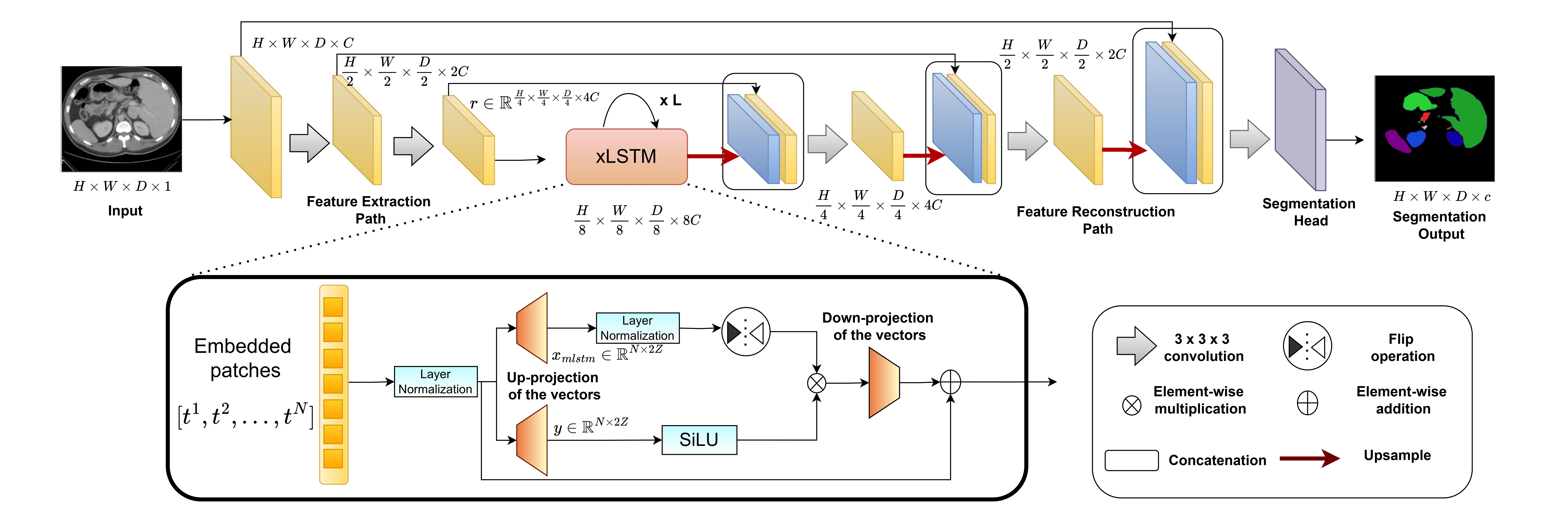
Architectural framework of UVixLSTM depicting the input processed by stacked layers of CNNs and Vision- xLSTM blocks. The intermediate feature representation from the feature extraction path is upsampled through the feature reconstruction path to obtain the final segmentation output.
Segmentation Results
Datasets
The Synapse dataset [3] comprises 30 CT volumes with sizes varying from 512 × 512 × 85 to 512 × 512 × 198. The CT volumes are annotated manually by domain experts to highlight the different abdominal organs. The model is trained to segment nine distinct organs of the abdominal cavity viz. spleen, left kidney, right kidney, liver, gall bladder, pancreas, stomach, right adrenal gland, and left adrenal gland. The spleen, liver, and stomach are classified as larger organs, whereas the kidneys, gall bladder, pancreas, and adrenal glands are smaller in size.
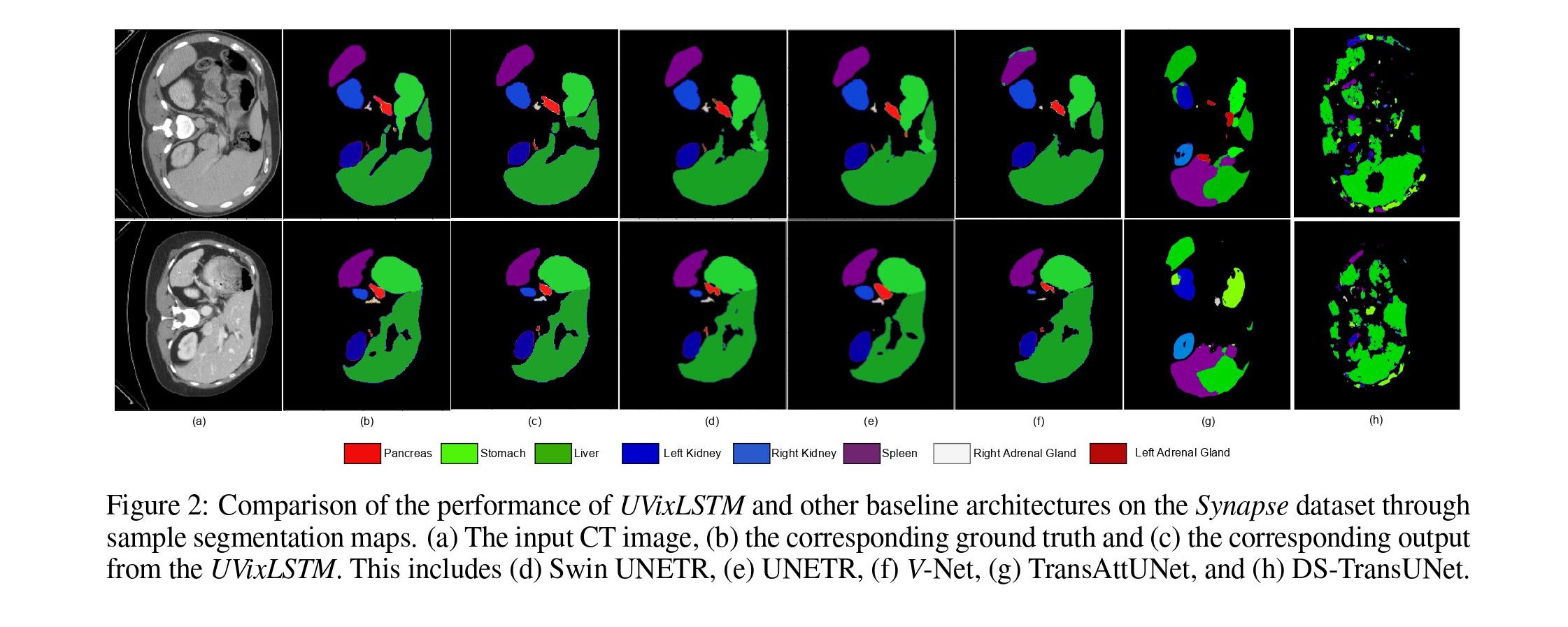
Qualitative Results
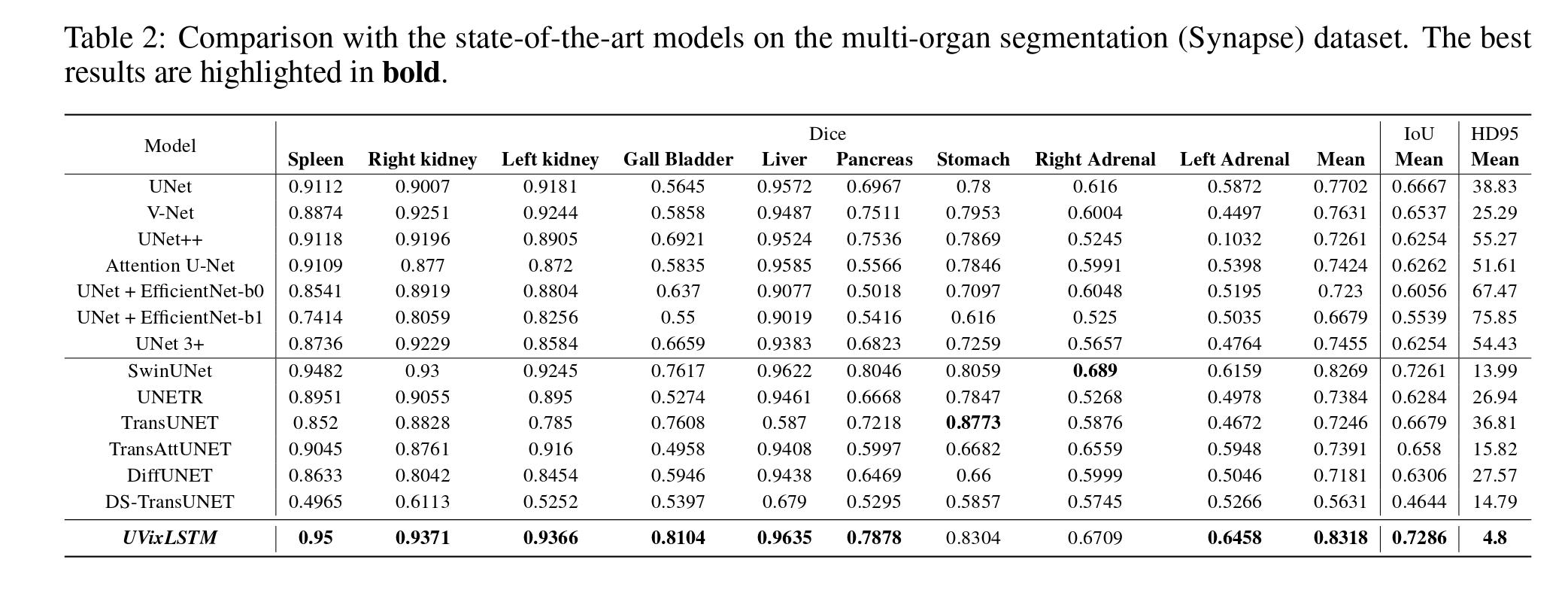
Quantitative Results
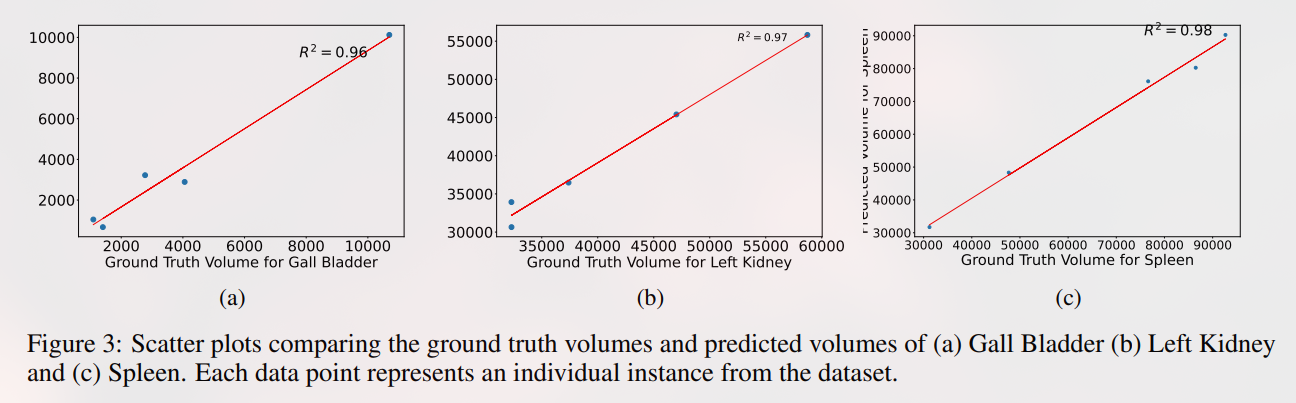
Regression Plots
Computational Complexity vs Flops
Citation
@article{dutta2024segmentation,
title={Are Vision xLSTM Embedded U-Nets More Reliable in Medical 3D Image Segmentation?},
author={Dutta, Pallabi and Bose, Soham and Roy, Swalpa Kumar and Mitra, Sushmita},
url={https://arxiv.org/abs/2406.16993},
journal={arXiv},
pp.={1-9},
year={2024}
}
References
- B. Alkin, M. Beck, and et al., “Vision-LSTM: xLSTM as generic vision backbone,” arXiv preprint arXiv:2406.04303, 2024.
- M. Beck, K. Pöppel, and et al., “xLSTM: Extended Long Short-Term Memory,” arXiv preprint arXiv:2405.04517, 2024.
- B. Landman, Z. Xu, and et al., “MICCAI multi-atlas labeling beyond the cranial vault–workshop and challenge,” in Proceedings of MICCAI Multi-Atlas Labeling beyond Cranial Vault—Workshop Challenge, p. 12, 2015.